Table of Contents¶
ALGO1 : Introduction à l'algorithmique¶
- Page du cours : https://perso.crans.org/besson/teach/info1_algo1_2019/
- Magistère d'Informatique de Rennes - ENS Rennes - Année 2019/2020
- Intervenants :
- Cours : Lilian Besson
- Travaux dirigés : Raphaël Truffet
- Références :
Cours Magistral 3¶
- Ce cours traite du paradigme "Diviser pour Régner",
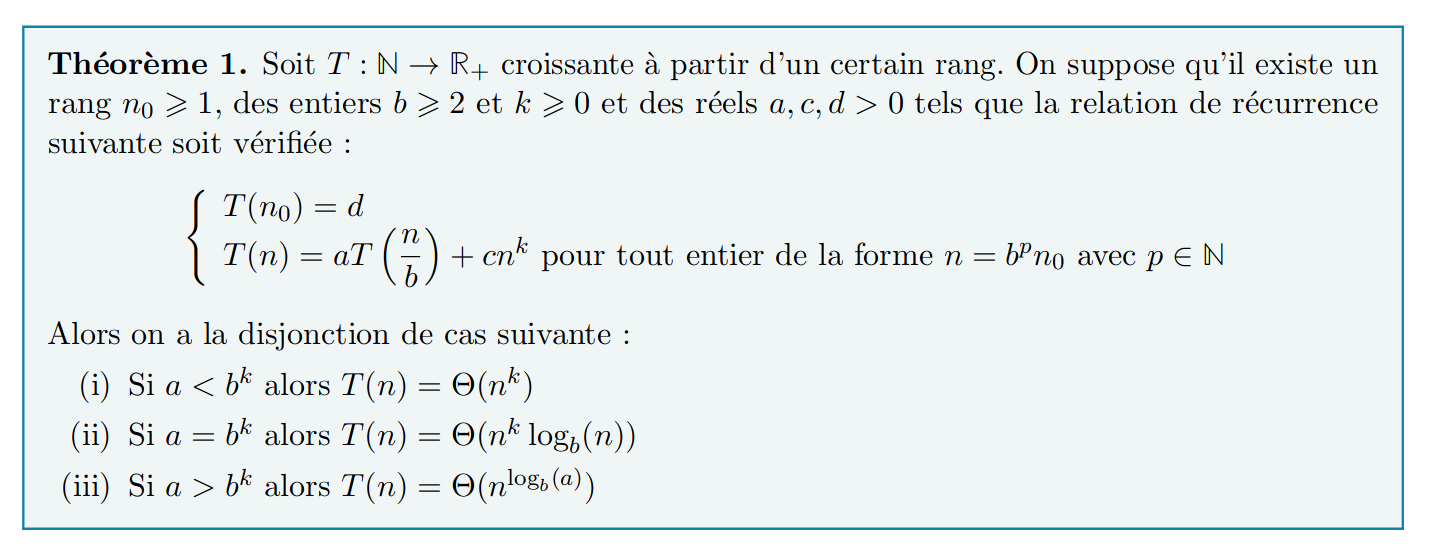
- Un important résultat théorique est donné (et prouvé) :
Ce notebook commence par donner quelques algorithmes "Diviser pour Régner" assez classiques, et permettent d'illustrer les deux premiers cas du "master theorem" (recherche dans une liste triée ou un tableau trié, tri fusion,
Ce théorème ne suffit pas à couvrir tous les différents algorithmes "Diviser pour Régner", avec comme contre exemple l'algorithme de tri rapide,
Puis on implémente les deux algorithmes présentés dans le cours, la multiplication de grands entiers par l'algorithme de Gauss-Karatsuba, et la multiplication de grandes matrices par l'algorithme de Strassen (Straßen).
Quelques algorithmes "diviser pour régner" classiques¶
Recherche dans un tableau trié en $\mathcal{O}(\log(n))$¶
Si un tableau T = [a1, ..., an] est trié, et qu'on donne une valeur x, on cherche un indice i tel que T[i] = ai = x, s'il existe (sans spécification s'il n'est pas unique), et une erreur si x n'est pas présent dans le tableau.
En utilisant des indices left et right, qu'on fait augmenter ou diminuer, on évite de faire des recopies du tableau.
La complexité de cet algorithme est en $\mathcal{O}(\log(n))$.
Avec le master theorem, on a $a=1, b=2, k=0$ : on divise les entrées en $a=2$ entrées de tailles $\leq b=2$ plus petites, sur lesquels on applique un traitement constant ($\mathcal{O}(n^{k=0})$ avant l'appel récursif (aucune recopie, juste des changements d'indices !).
def dichotomy_in_array(array, value, left=0, right=None):
if depth > len(array): raise KeyError
if right is None:
right = len(array) - 1
n = right - left + 1
if n == 0: raise KeyError
elif n == 1:
if array[left] == value:
return left
else: raise KeyError
index_of_middle = left + (n // 2)
middle_of_list = array[index_of_middle]
if value < middle_of_list: # search on left
return dichotomy_in_array(array, value, left=left, right=index_of_middle - 1)
elif value > middle_of_list: # search on right
return dichotomy_in_array(array, value, left=index_of_middle + 1, right=right)
else:
return index_of_middle
Et avec juste un peu d'affichage pour (un rappel ou pour) comprendre le fonctionnement :
def dichotomy_in_array(array, value, left=0, right=None, debug=False, depth=0):
if depth > len(array):
raise KeyError
if right is None:
right = len(array) - 1
n = right - left + 1
if debug: print(f" {' '*(depth+1)}Searching for {value} in sequence of size {n} = {array[left:right+1]}")
if n == 0:
raise KeyError
elif n == 1:
if array[left] == value:
return left
else:
raise KeyError
index_of_middle = left + (n // 2)
middle_of_list = array[index_of_middle]
#if debug: print(f" {' '*(depth+1)}{value} >/</=? {middle_of_list}")
if value < middle_of_list: # search on left
if debug: print(f" {' '*(depth+1)}-> going left...")
return dichotomy_in_array(array, value, left=left, right=index_of_middle - 1, debug=debug, depth=depth+1)
elif value > middle_of_list: # search on right
if debug: print(f" {' '*(depth+1)}-> going right...")
return dichotomy_in_array(array, value, left=index_of_middle + 1, right=right, debug=debug, depth=depth+1)
elif value == middle_of_list:
if debug: print(f" {' '*(depth+1)}-> found at index {index_of_middle} !")
return index_of_middle
else:
raise KeyError
Faisons quelques essais :
n = 16
example_of_array = list(range(n))
for value in example_of_array:
print(f"\n Looking for value {value} in {example_of_array} :")
dichotomy_in_array(example_of_array, value, debug=True)
Et maintenant des essais sur des entrées de tailles croissantes :
import random
def random_sorted_sequence(n, minint=0, maxint=1000):
sequence = [random.randint(minint, maxint) for _ in range(n)]
return sorted(sequence)
def test_dichotomy_in_array(n, debug=False, array=None):
if array is None:
array = random_sorted_sequence(n)
value = random.choice(array)
return dichotomy_in_array(array, value, debug=debug)
test_dichotomy_in_array(16, debug=True)
import sys
sys.setrecursionlimit(10000)
T = random_sorted_sequence(100)
%timeit test_dichotomy_in_array(100, array=T)
T = random_sorted_sequence(1000)
%timeit test_dichotomy_in_array(1000, array=T)
T = random_sorted_sequence(10000)
%timeit test_dichotomy_in_array(10000, array=T)
T = random_sorted_sequence(100000)
%timeit test_dichotomy_in_array(100000, array=T)
T = random_sorted_sequence(1000000)
%timeit test_dichotomy_in_array(1000000, array=T)
La complexité semble bien logarithmique en $n$ (ie., $\mathcal{O}(\log(n))$) !
Recherche dans une liste triée en $\mathcal{O}(n)$¶
C'est comme la recherche dans un tableau trié, sauf qu'on recopie la liste de gauche ou de droite lors des appels récursifs.
La complexité de cet algorithme est en $\mathcal{O}(n)$.
Avec le master theorem, on a $a=1, b=2, k=1$ : on divise les entrées en $a=2$ entrées de tailles $\leq b=2$ plus petites, sur lesquels on applique un traitement linéaire ($\mathcal{O}(n^{k=1})$ avant l'appel récursif (à cause des recopies !).
def dichotomy_in_list(sequence, value):
n = len(sequence)
if n == 0: raise KeyError
index_of_middle = n // 2
middle_of_list = sequence[index_of_middle]
if value < middle_of_list: # search on left
# creating this list takes O(n/2) time
left_list = sequence[:index_of_middle]
return dichotomy_in_list(left_list, value, debug=debug, depth=depth+1)
elif value > middle_of_list: # search on right
# creating this list takes O(n/2) time
right_list = sequence[index_of_middle:]
return index_of_middle + dichotomy_in_list(right_list, value, debug=debug, depth=depth+1)
else:
return index_of_middle
Et avec juste un peu d'affichage pour (un rappel ou pour) comprendre le fonctionnement :
def dichotomy_in_list(sequence, value, debug=False, depth=0):
n = len(sequence)
if debug:
print(f" {' '*(depth+1)}Sequence of size {n} = {sequence}")
if n == 0:
raise KeyError
index_of_middle = n // 2
middle_of_list = sequence[index_of_middle]
if value < middle_of_list: # search on left
left_list = sequence[:index_of_middle]
if debug: print(f" {' '*(depth+1)}-> going left, in {left_list} of size {len(left_list)}...")
return dichotomy_in_list(left_list, value, debug=debug, depth=depth+1)
elif value > middle_of_list: # search on right
right_list = sequence[index_of_middle:]
if debug: print(f" {' '*(depth+1)}-> going right, in {right_list} of size {len(right_list)}...")
return index_of_middle + dichotomy_in_list(right_list, value, debug=debug, depth=depth+1)
elif value == middle_of_list:
if debug: print(f" {' '*(depth+1)}-> found at index {index_of_middle} !")
return index_of_middle
else:
raise KeyError
Faisons quelques essais :
n = 16
example_of_list = list(range(n))
for value in example_of_list:
print(f"\n Looking for value {value} in {example_of_list} :")
dichotomy_in_list(example_of_list, value, debug=True)
Et maintenant des essais sur des entrées de tailles croissantes :
import random
def random_sorted_sequence(n, minint=0, maxint=1000):
sequence = [random.randint(minint, maxint) for _ in range(n)]
return sorted(sequence)
def test_dichotomy_in_list(n, debug=False, sequence=None):
if sequence is None:
sequence = random_sorted_sequence(n)
value = random.choice(sequence)
return dichotomy_in_list(sequence, value, debug=debug)
test_dichotomy_in_array(16, debug=True)
import sys
sys.setrecursionlimit(10000)
L = random_sorted_sequence(100)
%timeit test_dichotomy_in_list(100, sequence=L)
L = random_sorted_sequence(1000)
%timeit test_dichotomy_in_list(1000, sequence=L)
L = random_sorted_sequence(10000)
%timeit test_dichotomy_in_list(10000, sequence=L)
L = random_sorted_sequence(100000)
%timeit test_dichotomy_in_list(100000, sequence=L)
L = random_sorted_sequence(1000_000)
%timeit test_dichotomy_in_list(1000_000, sequence=L)
L = random_sorted_sequence(10_000_000)
%timeit test_dichotomy_in_list(10_000_000, sequence=L)
La complexité semble bien linéaire en $n$ (ie., $\mathcal{O}(n)$) !
Tri fusion (merge sort)¶
Problème :
- entrée = tableau
T = [a1, ..., an]denvaleurs - sortie = tableau trié par ordre croissant
- entrée = tableau
Algorithme :
- on divise en deux le tableau,
gauche = [a1, ..., a_n/2]etdroite = [a_n/2+1, ..., an], - on trie récursivement les deux sous tableaux,
- on fusionne (merge) les deux sous tableaux en un.
- on divise en deux le tableau,
La fusion est simple à réaliser : on commence tout à gauche des deux tableaux, on avance dans le tableau de gauche ou le tableau de droite, séquentiellement, jusqu'à avoir épuiser leurs valeurs, en prenant la valeur de gauche tant qu'elle est plus petite que celle de droite.
La complexité de cet algorithme est en $\mathcal{O}(n \log(n))$.
Avec le master theorem, on a $a=2, b=2, k=1$ : on divise les entrées en $a=2$ entrées de tailles $\leq b=2$ plus petites, sur lesquels on applique un traitement linéaire ($\mathcal{O}(n^{k=1})$ avant et après l'appel récursif.
def merge(left, right):
result = []
left_idx, right_idx = 0, 0 # growing pointers
while left_idx < len(left) and right_idx < len(right):
# this loop terminates because left_idx + right_idx is strictly increasing
# and bounded by len(left) + len(right)
# change the direction of this comparison to change the direction of the sort
if left[left_idx] <= right[right_idx]:
result.append(left[left_idx])
left_idx += 1
else:
result.append(right[right_idx])
right_idx += 1
# we still have values to take on the left
if left_idx < len(left):
result.extend(left[left_idx:])
# we still have values to take on the right
if right_idx < len(right):
result.extend(right[right_idx:])
return result
def merge_sort(m):
if len(m) <= 1:
return m
middle = len(m) // 2
# separating the array in two pieces is easy with Python
# but keep in mind, this takes a linear time to copy the arrays!
left = m[:middle]
right = m[middle:]
sorted_left = merge_sort(left)
sorted_right = merge_sort(right)
return list(merge(sorted_left, sorted_right))
Quelques tests :
L = random_sorted_sequence(100)
%timeit merge_sort(shuffle(L))
L = random_sorted_sequence(1000)
%timeit merge_sort(shuffle(L))
L = random_sorted_sequence(10_000)
%timeit merge_sort(shuffle(L))
L = random_sorted_sequence(100_000)
%timeit merge_sort(shuffle(L))
L = random_sorted_sequence(1000_000)
%timeit merge_sort(shuffle(L))
On pourrait vérifier que la complexité est bien en $\mathcal{O}(n \log(n))$.
import timeit
try:
from tqdm import tqdm_notebook as tqdm
except ImportError:
def tqdm(iterator, *args, **kwargs):
return iterator
values_n = np.array(np.floor(np.logspace(2, 6.5, num=50)), dtype=int)
values_L = [ random_sorted_sequence(n) for n in values_n ]
values_times = [
timeit.timeit(
stmt=f"merge_sort(shuffle({L}))",
number= 10000 if n <= 1000 else (1000 if n <= 10000 else (100 if n <= 100000 else 10)),
globals=globals()
)
for (n, L) in tqdm(list(zip(values_n, values_L)))
]
import numpy as np
import matplotlib as mpl
mpl.rcParams['figure.figsize'] = (8, 5)
mpl.rcParams['figure.dpi'] = 120
import matplotlib.pyplot as plt
import seaborn as sns
sns.set(context="notebook", style="whitegrid", palette="hls", font="sans-serif", font_scale=1.1)
plt.figure()
plt.xlabel("Size of the input array $n$")
plt.ylabel("Time in second")
plt.title("Time complexity of the merge sort algorithm (naive code in Python)")
plt.plot(values_n, values_times, "d-")
plt.show()
plt.figure()
plt.xlabel("Size of the input array $n$")
plt.ylabel("Time in milli-second, normalized by $n \log(n)$")
plt.title("Time complexity of the merge sort algorithm (naive code in Python)")
normalized_values_times = 1e6 * np.array(values_times) / (values_n * np.log(values_n))
min_time = 1e5
plt.plot(values_n[values_n >= min_time], normalized_values_times[values_n >= min_time], "d-")
plt.show()
Enveloppe convexe de points en 2D (quickhull)¶
Problème :
- entrée = tableau
T = [xy1, ..., xyn]denpoints dans le plan (xy = (x, y)) - sortie = tableau
hull = [xy_i1, ..., xy_ip]desppoints constituant l'enveloppe convexe desnpoints d'entrée
- entrée = tableau
Algorithme :
- on identifie le point le plus en bas à gauche $P_{bg}$, le point le plus en haut à droite $P_{hd}$,
- la diagonale $D$ est le segment orienté qui va de $P_{bg}$ à $P_{hd}$,
- on sépare l'ensemble en deux, les points à gauche de $D$ (coin bas droite), ceux à droite de $D$,
- on calcule les deux enveloppes convexes $E_g$ et $E_d$ des deux ensembles de points (récursivement),
- on fusionne les deux enveloppes convexes $E_g$ et $E_d$ en les ordonnant comme il faut (et en enlevant l'arête $P_{hd} \to P_{pg}$ qui est présente dans $E_g$ et $P_{pg} \to P_{hd}$ présente dans $E_d$.
La fusion est naïve simple à réaliser. Le test pour savoir si un point est à gauche, à droite, ou sur $D$, se fait en temps $O(1)$ avec un calcul de produit scalaire.
exemple_points = [(1, 3) for _ in range(21)]
for i in range(1, 21):
x, y = exemple_points[i-1]
exemple_points[i] = (x * 17) % 23, (y * 17) % 19
plt.figure()
plt.title("Des points en 2D")
Xs = [p[0] for p in exemple_points]
Ys = [p[1] for p in exemple_points]
ax = plt.scatter(Xs, Ys)
plt.show()
def alpha(a, b, c):
xa, ya = a
xb, yb = b
xc, yc = c
return (xb - xa) * (yc - ya) - (xc - xa) * (yb - ya)
p = exemple_points
for i in range(2, 21):
for j in range(1, i):
for k in range(0, j):
if alpha(p[i], p[j], p[k]) == 0:
print(f"Les trois points #{i}, #{j} et #{k} sont alignés ({p[i]}, {p[j]} et {p[k]}).")
plt.figure()
plt.title("Des points en 2D, certains sont alignés")
Xs = [p[0] for p in exemple_points]
Ys = [p[1] for p in exemple_points]
plt.scatter(Xs, Ys)
p = exemple_points
for i in range(2, 21):
for j in range(1, i):
for k in range(0, j):
if alpha(p[i], p[j], p[k]) == 0:
plt.plot([p[i][0], p[j][0], p[k][0]], [p[i][1], p[j][1], p[k][1]], ':')
plt.show()
def plus_bas(points):
""" Trouve le point (xa, ya) le plus en bas à gauche, en temps linéaire."""
n = len(points)
xa, ya = points[0]
for j in range(1, n):
xj, yj = points[j]
if (ya > yj) or (ya == yj and xa > xj):
xa, ya = xj, yj
return xa, ya
plus_bas(exemple_points) # (12, 2)
def plus_haut(points):
""" Trouve le point (xb, yb) le plus en haut à droite, en temps linéaire."""
n = len(points)
xb, yb = points[0]
for j in range(1, n):
xj, yj = points[j]
if (yb < yj) or (yb == yj and xb < xj):
xb, yb = xj, yj
return xb, yb
plus_haut(exemple_points) # (21, 8)
def plus_a_droite(b, h, points):
if not points:
return b, []
t = points[0]
q = points[1:]
m, d = plus_a_droite(b, h, q) # recursif !
angle = alpha(b, t, h)
if angle <= 0:
return m, d
else:
if not d:
return t, [t]
if angle > alpha(b, m, h):
return t, [t] + d
else:
return m, [t] + d
def quickHull_droite(b, h, points):
m, d = plus_a_droite(b, h, points)
# si aucun point n'est à droite du segment orienté [b h], on en garde un seul, [b]
if not d:
return [b]
# on fait les deux appels récursifs !
return quickHull_droite(b, m, d) + quickHull_droite(m, h, d)
def quickHull(points):
b = plus_haut(points)
h = plus_bas(points)
return quickHull_droite(b, h, points) + quickHull_droite(h, b, points)
quickHull(exemple_points)
La complexité de cet algorithme est en $\mathcal{O}(n \log(n))$.
Avec le master theorem, on a $a=2, b=2, k=1$ : on divise les entrées en $a=2$ entrées de tailles $\leq b=2$ plus petites, sur lesquels on applique un traitement linéaire ($\mathcal{O}(n^{k=1})$ avant et après l'appel récursif. (En supposant les points uniformément répartis dans le plan.)
On pourrait l'implémenter, et vérifier la complexité sur des entrées aléatoires (ie. les points sont tirés uniformément dans une boîte carrée $(x_i,y_i) \sim [x_\min,x_\max] \times [y_\min,y_\max]$).
Attention ici avec Python, les ajout en tête de listes peuvent prendre un coût linéaire (et pas constant comme en OCaml) et les concaténations de listes prennent un coût linéaire ! Donc la vraie implémentation sera plus coûteuse que ce $\mathcal{O}(n \log(n))$ annoncé.
Un exemple¶
plt.figure()
plt.title("Des points en 2D, et leur enveloppe convexe")
Xs = [p[0] for p in exemple_points]
Ys = [p[1] for p in exemple_points]
plt.scatter(Xs, Ys)
p = exemple_points
enveloppe = quickHull(p)
q = enveloppe
h = len(enveloppe)
for i in range(h - 1):
plt.plot([q[i][0], q[i+1][0]], [q[i][1], q[i+1][1]])
plt.plot([q[-1][0], q[0][0]], [q[-1][1], q[0][1]])
plt.show()
Exemples aléatoires de taille contrôlée¶
import random
def points_aleatoires(n=100, xmin=-10, xmax=10, ymin=-10, ymax=10):
return [(random.randint(xmin, xmax), random.randint(ymin, ymax)) for _ in range(n)]
xmin, xmax, ymin, ymax = -10, 10, -10, 10
for n in [5, 10, 50, 100, 500, 1000]:
p = points_aleatoires(n=n, xmin=xmin, xmax=xmax, ymin=ymin, ymax=ymax)
_ = plt.figure()
_ = plt.title(f"{n} points aléatoires dans [{xmin},{xmax}] x [{ymin},{ymax}] en 2D, et leur enveloppe convexe")
Xs = [pi[0] for pi in p]
Ys = [pi[1] for pi in p]
_ = plt.scatter(Xs, Ys)
enveloppe = quickHull(p)
q = enveloppe
h = len(enveloppe)
for i in range(h - 1):
_ = plt.plot([q[i][0], q[i+1][0]], [q[i][1], q[i+1][1]])
_ = plt.plot([q[-1][0], q[0][0]], [q[-1][1], q[0][1]])
_ = plt.show()
Complexité temporelle de ce calcul d'enveloppe convexe¶
values_n = np.array(np.floor(np.logspace(1, 3.5, num=20)), dtype=int)
values_points = [ points_aleatoires(n=n) for n in values_n ]
values_times = [
timeit.timeit(
stmt=f"quickHull({points})",
number= 1000 if n <= 1000 else (100 if n <= 10000 else (10 if n <= 100000 else 1)),
globals=globals()
)
for (n, points) in tqdm(list(zip(values_n, values_points)))
]
plt.figure()
plt.xlabel("Size of the input array $n$")
plt.ylabel("Time in second")
plt.title("Time complexity of the quick hull algorithm (naive code in Python)")
plt.plot(values_n, values_times, "d-")
plt.show()
plt.figure()
plt.xlabel("Size of the input array $n$")
plt.ylabel("Time in milli-second, normalized by $n \log(n)$")
plt.title("Time complexity of the quick hull algorithm (naive code in Python)")
normalized_values_times = 1e6 * np.array(values_times) / (values_n * np.log(values_n))
min_time = 1
plt.plot(values_n[values_n >= min_time], normalized_values_times[values_n >= min_time], "d-")
plt.show()
Algorithme de Gauss-Karatsuba¶
Problème :
- entrée = deux nombres
xety, avecnchiffres dans leurs écritures décimales, - sortie = un nombre
z = x * yproduit des deux nombres.
- entrée = deux nombres
Algorithme :
- on découpe les deux nombres en nombres avec au plus
n/2chiffres :- découper
x=b + 10^{n/2} a, avecaetbde taille au plusn/2 - découper
y=d + 10^{n/2} c, aveccetdde taille au plusn/2
- découper
- trois appels récursifs au produit de nombres, de tailles deux fois plus petites
- on calcule
a * cetb * d - l'astuce vient de
ad_plus_bc = ad + bc = (a+b)(c+d) - ac - bdcomme(a+b)(c+d) = ac + ad + bc + bd x y = (b + 10^{n/2} a) (d + 10^{n/2} c) = bd + 10^{n/2} (ad_plus_bc) + 10^{n} (a c)
- on découpe les deux nombres en nombres avec au plus
La complexité de cet algorithme est en $\mathcal{O}(n^{\log_2(3)})$, asymptotiquement meilleur que $\mathcal{O}(n^2)$ la méthode naïve.
Avec le master theorem, on a $a=3, b=2, k=1$ : on divise les entrées en $a=7$ entrées de tailles $\leq b=2$ plus petites [1], sur lesquels on applique un traitement linéaire (toutes les additions $\mathcal{O}(n^{k=1})$ avant et après l'appel récursif.
Donc $a = 3 > b^k = 2$ ce qui donne $T(n) = \mathcal{O}(n^{\log_b(a)})$.
En comparaison, la méthode naïve, que voici, sera en avec $a=4$ appels récursifs, soit $a = 4 > b^k = 2$ ce qui donne $T(n) = \mathcal{O}(n^{\log_b(a)}) = \mathcal{O}(n^2)$.
def naivemult(x, y, base=10):
""" Function to multiply 2 numbers using the grade school algorithm."""
if len(str(x)) == 1 or len(str(y)) == 1:
return x*y
else:
n = max(len(str(x)),len(str(y)))
# that's suboptimal, and ugly, but it's quick to write
nby2 = n // 2
# split x in b + 10^{n/2} a, with a and b of sizes at most n/2
a = x // base**(nby2)
b = x % base**(nby2)
# split y in d + 10^{n/2} a, with c and d of sizes at most n/2
c = y // base**(nby2)
d = y % base**(nby2)
# we make 3 calls to entries which are 2 times smaller
ac = naivemult(a, c)
ad = naivemult(a, d)
bd = naivemult(b, d)
bc = naivemult(b, c)
# x y = (b + 10^{n/2} a) (d + 10^{n/2} c)
# ==> x y = bd + 10^{n/2} (b c + a d) + 10^{n} (a c)
# this little trick, writing n as 2*nby2 takes care of both even and odd n
prod = ac * base**(2*nby2) + ((ad + bc) * base**nby2) + bd
return prod
Et la fonction implémentant l'algorithme de Karatsuba.
def karatsuba(x, y, base=10):
""" Function to multiply 2 numbers in a more efficient manner than the grade school algorithm."""
if len(str(x)) == 1 or len(str(y)) == 1:
return x*y
else:
n = max(len(str(x)),len(str(y)))
# that's suboptimal, and ugly, but it's quick to write
nby2 = n // 2
# split x in b + 10^{n/2} a, with a and b of sizes at most n/2
a = x // base**(nby2)
b = x % base**(nby2)
# split y in d + 10^{n/2} c, with c and d of sizes at most n/2
c = y // base**(nby2)
d = y % base**(nby2)
# we make 3 calls to entries which are 2 times smaller
ac = karatsuba(a, c)
bd = karatsuba(b, d)
# ad + bc = (a+b)(c+d) - ac - bd as (a+b)(c+d) = ac + ad + bc + bd
ad_plus_bc = karatsuba(a + b, c + d) - ac - bd
# x y = (b + 10^{n/2} a) (d + 10^{n/2} c)
# ==> x y = bd + 10^{n/2} (ad_plus_bc) + 10^{n} (a c)
# this little trick, writing n as 2*nby2 takes care of both even and odd n
prod = ac * base**(2*nby2) + (ad_plus_bc * base**nby2) + bd
return prod
Un exemple :
x = 1234
y = 4567
x * y
naivemult(x, y)
karatsuba(x, y)
Des exemples de grandes tailles :
def rand_largeint(n=1024):
return int("".join(str(random.randint(0, 9)) for _ in range(n)))
x = rand_largeint(1024)
y = rand_largeint(1024)
%timeit x * y
%timeit naivemult(x, y)
%timeit karatsuba(x, y)
Quelle est l'influence de la base ?
n = 1024
base = 10
%timeit rand_largeint(n) * rand_largeint(n)
%timeit naivemult(rand_largeint(n), rand_largeint(n), base=base)
%timeit karatsuba(rand_largeint(n), rand_largeint(n), base=base)
n = 1024
base = 2
%timeit rand_largeint(n) * rand_largeint(n)
%timeit naivemult(rand_largeint(n), rand_largeint(n), base=base)
%timeit karatsuba(rand_largeint(n), rand_largeint(n), base=base)
Et pour des entrées de tailles croissantes :
for n in [2**3, 2**4, 2**5, 2**6, 2**7, 2**8, 2**9, 2**10, 2**11, 2**12]:
print(f"\nFor n = {n} : Python native, naive then Karatsuba :")
x = rand_largeint(n)
y = rand_largeint(n)
%timeit x * y # crazy fast!
%timeit naivemult(x, y)
%timeit karatsuba(x, y)
Ce n'est pas facile de vérifier le comportement en $n^{\log_2(3)}$ mais on voit déjà que la méthode de Gauss-Karatsuba est bien plus rapide que la méthode naïve !
Algorithme de Strassen¶
- On cherche à multiplier deux matrices $A, B \in\mathbb{K}^{n \times n}$, où $\mathbb{K}$ est un anneau commutatif quelconque (e.g., les entiers, ou les rationnels, ou les flottants), et $n\in\mathbb{N}$ est un entier.
On va rappeler un algorithme naïf (de complexité temporelle asymptotque en $\Theta(n^3)$), un algorithme récursif mais qui n'est pas plus efficace que l'algorithme naïf, et un algorithme récursif qui attend une complexité asymptotique plus effaice ($\Theta(n^{\log_2(7)})$).
Cf le cours pour l'algorithme, ou https://fr.wikipedia.org/wiki/Algorithme_de_Strassen.
Part of this code is coming from this blog post.
Méthode naïve, "méthode i k j"¶
On écrit $C = A \times B$, et on rappelle que $\forall i, j \in[n], C_{i,j} = \sum_{k=1}^n A_{i,k} B_{k,j}$.
Donc on peut calculer cela avec trois boucles for imbriquées, pour $i, k, j$.
On va utiliser des tableaux numpy et pas des listes de listes, pour plus de simplicités.
import numpy as np
Attention : pour ne pas être trop lent, j'utilise la compilation "just in time" proposée par le projet numba, pour faire que ces fonctions naïves soient aussi efficaces que celles de Numpy (en gros).
try:
from numba import jit
except ImportError:
def jit(f, *args, **kwargs):
return f
@jit
def ikjMatrixProduct(A, B):
""" Produit matriciel naïf, en O(n^3) opérations."""
n = len(A)
C = np.zeros((n, n), dtype=type(A[0,0]))
for i in range(n):
for k in range(n):
for j in range(n):
C[i,j] += A[i,k] * B[k,j]
return C
Méthode naïve récursive¶
On va d'abord définir nos propres opérations d'additions et de soustractions de matrices dans $\mathbb{K}^{n \times n}$.
@jit
def add(A, B):
n = len(A)
C = np.zeros((n, n), dtype=type(A[0,0]))
for i in range(n):
for j in range(n):
C[i,j] = A[i,j] + B[i,j]
return C
@jit
def subtract(A, B):
n = len(A)
C = np.zeros((n, n), dtype=type(A[0,0]))
for i in range(n):
for j in range(n):
C[i,j] = A[i,j] - B[i,j]
return C
La méthode récursive mais naïve est la suivante. On découpe les matrices de taille $n \times n$ en quatre matrices de tailles $(n/2) \times (n/2)$. $$ A B = \begin{array}{l} \begin{pmatrix} a_{11} & a_{12}\\ a_{21} & a_{22} \end{pmatrix} \end{array} \begin{array}{l} \begin{pmatrix} b_{11} & b_{12}\\ b_{21} & b_{22} \end{pmatrix}
\end{array}¶
C = \begin{array}{l} \begin{pmatrix} c_{11} = p_1 + p2 = a_{11}b_{11} + a_{12}b_{21} & c_{12} = p_3 + p4 = a_{11}b_{12} + a_{12}b_{22}\\ c_{21} = p_5 + p6 = a_{21}b_{11} + a_{22}b_{21} & c_{22} = p_7 + p8 = a_{21}b_{12} + a_{22}b_{22} \end{pmatrix} \end{array} $$
def naive_recursive(A, B, leaf_size=LEAF_SIZE):
n = len(A)
if n <= leaf_size:
return ikjMatrixProduct(A, B)
else:
# initializing the new sub-matrices
newSize = n//2
a11 = A[:newSize, :newSize] # top left
a12 = A[:newSize, newSize:] # top right
a21 = A[newSize:, :newSize] # bottom left
a22 = A[newSize:, newSize:] # bottom right
b11 = B[:newSize, :newSize] # top left
b12 = B[:newSize, newSize:] # top right
b21 = B[newSize:, :newSize] # bottom left
b22 = B[newSize:, newSize:] # bottom right
# Calculating p1 to p8:
p1 = naive_recursive(a11, b11, leaf_size=leaf_size)
p2 = naive_recursive(a12, b21, leaf_size=leaf_size)
p3 = naive_recursive(a11, b12, leaf_size=leaf_size)
p4 = naive_recursive(a12, b22, leaf_size=leaf_size)
p5 = naive_recursive(a21, b11, leaf_size=leaf_size)
p6 = naive_recursive(a22, b21, leaf_size=leaf_size)
p7 = naive_recursive(a21, b12, leaf_size=leaf_size)
p8 = naive_recursive(a22, b22, leaf_size=leaf_size)
# calculating c11, c21, c11 and c22:
c11 = add(p1, p2)
c12 = add(p3, p4)
c21 = add(p5, p6)
c22 = add(p7, p8)
# Grouping the results obtained in a single matrix:
C = np.zeros((n, n), dtype=type(A[0,0]))
for i in range(newSize):
for j in range(newSize):
C[i,j] = c11[i,j]
C[i,j + newSize] = c12[i,j]
C[i + newSize,j] = c21[i,j]
C[i + newSize,j + newSize] = c22[i,j]
return C
La complexité de cet algorithme est en $\mathcal{O}(n^{\log_2(8)}) = \mathcal{O}(n^3)$ équivalent à la méthode naïve non récursive.
Avec le master theorem, on a $a=8, b=2, k=2$ : on divise les entrées en $8$ entrées de tailles plus petites, et on fait $a=8$ calculs sur des entrées de tailles $\leq b=2$ plus petites [1], sur lesquels on applique un traitement quadratique (toutes les additions $\mathcal{O}(n^{k=2})$ avant et après l'appel récursif.
Donc $a = 8 > b^k = 4$ ce qui donne $T(n) = \mathcal{O}(n^{\log_b(a)})$.
[1] : c'est $n$ qui est divisé par deux, bien que $n^2$ soit divisé par $4$ et que l'on puisse considérer que la taille d'une matrice $(n,m)$ est $n \times m$.
def naive(A, B, leaf_size=LEAF_SIZE):
assert isinstance(A, np.ndarray) and isinstance(B, np.ndarray)
assert len(A) == len(A[0]) == len(B) == len(B[0])
# Make the matrices bigger so that you can apply the strassen
# algorithm recursively without having to deal with odd matrix sizes
nextPowerOfTwo = lambda n: 2**int(ceil(log(n,2)))
n = len(A)
m = nextPowerOfTwo(n)
APrep = np.zeros((m, m), dtype=type(A[0,0]))
BPrep = np.zeros((m, m), dtype=type(A[0,0]))
for i in range(n):
for j in range(n):
APrep[i,j] = A[i,j]
BPrep[i,j] = B[i,j]
CPrep = naive_recursive(APrep, BPrep, leaf_size=leaf_size)
C = np.zeros((n, n), dtype=type(A[0,0]))
for i in range(n):
for j in range(n):
C[i,j] = CPrep[i,j]
return C
Méthode récursive de Strassen¶
La méthode récursive de Strassen est la suivante. On découpe les matrices de taille $n \times n$ en quatre matrices de tailles $(n/2) \times (n/2)$. $$ A B = \begin{array}{l} \begin{pmatrix} a_{11} & a_{12}\\ a_{21} & a_{22} \end{pmatrix} \end{array} \begin{array}{l} \begin{pmatrix} b_{11} & b_{12}\\ b_{21} & b_{22} \end{pmatrix}
\end{array}¶
C = \begin{array}{l} \begin{pmatrix} c_{11} = p1 + p4 - p5 + p7 & c_{12} = p3 + p5 \\ c_{21} = p2 + p4 & c_{22} = p1 + p3 - p2 + p6 \\ \end{pmatrix} \end{array} $$ Avec les 7 produits intermédiaires suivants : $$ p1 = (a{11}+a{22}) (b{11}+b{22}) \ p2 = (a{21}+a{22}) (b{11}) \ p3 = (a{11}) (b{12} - b{22}) \ p4 = (a_{22}) (b{21} - b{11}) \ p5 = (a{11}+a{12}) (b{22}) \ p6 = (a{21}-a_{11}) (b{11}+b{12}) \ p7 = (a{12}-a{22}) * (b{21}+b{22}) $$ On fait plus d'additions mais moins de produits, et les additions sont en $\Theta(n^2)$, les produits sont plus couteux, donc on va gagner (asymptotiquement) !
La complexité de cet algorithme est en $\mathcal{O}(n^{\log_2(7)})$, asymptotiquement meilleur que $\mathcal{O}(n^3)$ la méthode naïve.
Avec le master theorem, on a $a=7, b=2, k=2$ : on divise les entrées en $8$ entrées de tailles 2 fois plus petites, mais on ne fait que $a=7$ calculs sur entrées de tailles $\leq b=2$ plus petites [1], sur lesquels on applique un traitement quadratique (toutes les additions $\mathcal{O}(n^{k=2})$ avant et après l'appel récursif.
Donc $a = 7 > b^k = 4$ ce qui donne $T(n) = \mathcal{O}(n^{\log_b(a)})$.
[1] : c'est $n$ qui est divisé par deux, bien que $n^2$ soit divisé par $4$ et que l'on puisse considérer que la taille d'une matrice $(n,m)$ est $n \times m$.
On va encore utiliser cette idée de leaf_size : dès que les entrées sont trop petites, on réutilise l'algorithme naïf.
LEAF_SIZE = 64
def strassenR(A, B, leaf_size=LEAF_SIZE):
n = len(A)
if n <= leaf_size:
return ikjMatrixProduct(A, B)
else:
# initializing the new sub-matrices
newSize = n//2
a11 = A[:newSize, :newSize] # top left
a12 = A[:newSize, newSize:] # top right
a21 = A[newSize:, :newSize] # bottom left
a22 = A[newSize:, newSize:] # bottom right
b11 = B[:newSize, :newSize] # top left
b12 = B[:newSize, newSize:] # top right
b21 = B[newSize:, :newSize] # bottom left
b22 = B[newSize:, newSize:] # bottom right
# Calculating p1 to p7:
p1 = strassenR(add(a11, a22), add(b11, b22), leaf_size=leaf_size) # p1 = (a11+a22) * (b11+b22)
p2 = strassenR(add(a21, a22), b11, leaf_size=leaf_size) # p2 = (a21+a22) * (b11)
p3 = strassenR(a11, subtract(b12, b22), leaf_size=leaf_size) # p3 = (a11) * (b12 - b22)
p4 = strassenR(a22, subtract(b21, b11), leaf_size=leaf_size) # p4 = (a22) * (b21 - b11)
p5 = strassenR(add(a11, a12), b22, leaf_size=leaf_size) # p5 = (a11+a12) * (b22)
p6 = strassenR(subtract(a21, a11), add(b11, b12), leaf_size=leaf_size) # p6 = (a21-a11) * (b11+b12)
p7 = strassenR(subtract(a12, a22), add(b21, b22), leaf_size=leaf_size) # p7 = (a12-a22) * (b21+b22)
# calculating c21, c21, c11 e c22:
c12 = add(p3, p5) # c12 = p3 + p5
c21 = add(p2, p4) # c21 = p2 + p4
c11 = subtract(add(add(p1, p4), p7), p5) # c11 = p1 + p4 - p5 + p7
c22 = subtract(add(add(p1, p3), p6), p2) # c22 = p1 + p3 - p2 + p6
# Grouping the results obtained in a single matrix:
C = np.zeros((n, n), dtype=type(A[0,0]))
for i in range(newSize):
for j in range(newSize):
C[i,j] = c11[i,j]
C[i,j + newSize] = c12[i,j]
C[i + newSize,j] = c21[i,j]
C[i + newSize,j + newSize] = c22[i,j]
return C
def strassen(A, B, leaf_size=LEAF_SIZE):
assert isinstance(A, np.ndarray) and isinstance(B, np.ndarray)
assert len(A) == len(A[0]) == len(B) == len(B[0])
# Make the matrices bigger so that you can apply the strassen
# algorithm recursively without having to deal with odd matrix sizes
nextPowerOfTwo = lambda n: 2**int(ceil(log(n,2)))
n = len(A)
m = nextPowerOfTwo(n)
APrep = np.zeros((m, m), dtype=type(A[0,0]))
BPrep = np.zeros((m, m), dtype=type(A[0,0]))
for i in range(n):
for j in range(n):
APrep[i,j] = A[i,j]
BPrep[i,j] = B[i,j]
CPrep = strassenR(APrep, BPrep, leaf_size=leaf_size)
C = np.zeros((n, n), dtype=type(A[0,0]))
for i in range(n):
for j in range(n):
C[i,j] = CPrep[i,j]
return C
En fait, on devrait essayer d'utiliser les opérations les plus efficaces pour les additions et soustractions de matrices, afin de vraiment voir où se situe le gain de l'algorithme de Strassen.
def strassenR_with_numpy_for_add_sub(A, B, leaf_size=LEAF_SIZE):
n = len(A)
if n <= leaf_size:
return ikjMatrixProduct(A, B)
else:
# initializing the new sub-matrices
newSize = n//2
# dividing the matrices in 4 sub-matrices:
a11 = A[:newSize, :newSize] # top left
a12 = A[:newSize, newSize:] # top right
a21 = A[newSize:, :newSize] # bottom left
a22 = A[newSize:, newSize:] # bottom right
b11 = B[:newSize, :newSize] # top left
b12 = B[:newSize, newSize:] # top right
b21 = B[newSize:, :newSize] # bottom left
b22 = B[newSize:, newSize:] # bottom right
# Calculating p1 to p7:
p1 = strassenR_with_numpy_for_add_sub(a11 + a22, b11 + b22, leaf_size=leaf_size) # p1 = (a11+a22) * (b11+b22)
p2 = strassenR_with_numpy_for_add_sub(a21 + a22, b11, leaf_size=leaf_size) # p2 = (a21+a22) * (b11)
p3 = strassenR_with_numpy_for_add_sub(a11, b12 - b22, leaf_size=leaf_size) # p3 = (a11) * (b12 - b22)
p4 = strassenR_with_numpy_for_add_sub(a22, b21 - b11, leaf_size=leaf_size) # p4 = (a22) * (b21 - b11)
p5 = strassenR_with_numpy_for_add_sub(a11 + a12, b22, leaf_size=leaf_size) # p5 = (a11+a12) * (b22)
p6 = strassenR_with_numpy_for_add_sub(a21 - a11, b11 + b12, leaf_size=leaf_size) # p6 = (a21-a11) * (b11+b12)
p7 = strassenR_with_numpy_for_add_sub(a12 - a22, b21 + b22, leaf_size=leaf_size) # p7 = (a12-a22) * (b21+b22)
# calculating c21, c21, c11 e c22:
c12 = p3 + p5 # c12 = p3 + p5
c21 = p2 + p4 # c21 = p2 + p4
c11 = p1 + p4 + p7 - p5 # c11 = p1 + p4 - p5 + p7
c22 = p1 + p3 + p6 - p2 # c22 = p1 + p3 - p2 + p6
# Grouping the results obtained in a single matrix:
C = np.zeros((n, n), dtype=type(A[0,0]))
C[:newSize, :newSize] = c11
C[:newSize, newSize:] = c12
C[newSize:, :newSize] = c21
C[newSize:, newSize:] = c22
return C
def strassen_with_numpy_for_add_sub(A, B, leaf_size=LEAF_SIZE):
assert isinstance(A, np.ndarray) and isinstance(B, np.ndarray)
assert len(A) == len(A[0]) == len(B) == len(B[0])
# Make the matrices bigger so that you can apply the strassen
# algorithm recursively without having to deal with odd matrix sizes
nextPowerOfTwo = lambda n: 2**int(ceil(log(n,2)))
n = len(A)
m = nextPowerOfTwo(n)
APrep = np.zeros((m, m), dtype=type(A[0,0]))
BPrep = np.zeros((m, m), dtype=type(A[0,0]))
APrep[:n, :n] = A
BPrep[:n, :n] = B
CPrep = strassenR_with_numpy_for_add_sub(APrep, BPrep, leaf_size=leaf_size)
return CPrep[:n, :n]
Pour générer des exemples de tailles grandissantes :
import random
def random_matrix(n, minint=0, maxint=1000):
A = np.zeros((n, n), dtype=int)
for i in range(n):
for j in range(n):
A[i, j] = random.randint(minint, maxint)
return A
A = random_matrix(4)
A
B = random_matrix(4)
B
On vérifie sur un exemple que nos deux algorithmes calculant le produit $AB$ de matrices sont corrects :
A @ B
ikjMatrixProduct(A, B)
strassenR(A, B, leaf_size=1)
strassenR(A, B, leaf_size=2)
strassenR_with_numpy_for_add_sub(A, B, leaf_size=1)
strassenR_with_numpy_for_add_sub(A, B, leaf_size=2)
La première version n'utilisait pas la compilation "just in time" offerte par Numba, et c'était très lent ! Il y avait aussi beaucoup trop d'allocations mémoire !
J'ai optimisé et (naïvement) simplifié le code, en enlevant les déclarations mémoires inutiles, mais surtout en utilisant numba.jit pour optimiser les opérations add et substract, et on obtient des temps bien plus courts :
for n in tqdm([2**5, 2**6, 2**7, 2**8, 2**9]):
print(f"\nFor n = {n} : numpy, ten ikj naive algorithm, then Strassen, then (slightly) faster Strassen :")
A = random_matrix(n)
B = random_matrix(n)
C = A @ B
%timeit A @ B # crazy fast!
assert np.all(np.isclose(ikjMatrixProduct(A, B), C))
%timeit ikjMatrixProduct(A, B)
assert np.all(np.isclose(strassen(A, B, leaf_size=4), C))
%timeit strassen(A, B, leaf_size=4)
assert np.all(np.isclose(strassen_with_numpy_for_add_sub(A, B, leaf_size=4), C))
%timeit strassen_with_numpy_for_add_sub(A, B, leaf_size=4)
Est-ce que les temps sont vraiment différents si les deux matrices A et B changent à chaque test ? Pas vraiment ! Mais pour les petites valeurs de n, les quatre façons de multiplier sont toutes aussi lentes : le temps est principalement passé à générer A et B !
for n in tqdm([2**5, 2**6, 2**7, 2**8, 2**9, 2**10]):
print(f"\nFor n = {n} : numpy, ten ikj naive algorithm, then Strassen, then (slightly) faster Strassen :")
%timeit random_matrix(n) @ random_matrix(n)
%timeit ikjMatrixProduct(random_matrix(n), random_matrix(n))
%timeit strassen(random_matrix(n), random_matrix(n), leaf_size=64)
%timeit strassen_with_numpy_for_add_sub(random_matrix(n), random_matrix(n), leaf_size=64)
for n in tqdm([2**7, 2**8, 2**9]):
print(f"\nFor n = {n} : numpy, ten ikj naive algorithm, then Strassen, then (slightly) faster Strassen :")
%timeit random_matrix(n) @ random_matrix(n)
%timeit ikjMatrixProduct(random_matrix(n), random_matrix(n))
for leaf_size in tqdm([8, 32, 64, 128]):
print(f"Both Strassen, with a leaf size = {leaf_size}")
%timeit strassen(random_matrix(n), random_matrix(n), leaf_size=leaf_size)
%timeit strassen_with_numpy_for_add_sub(random_matrix(n), random_matrix(n), leaf_size=leaf_size)
Transformée de Fourier Discrète (DFT), implémentation naïve¶
Ce premier algorithme est simple à écrire, et a une complexité en $\Theta(n^2)$.
def dft(x):
n = len(x)
omega = lambda j, k: np.exp(- (j * k) * 2j * np.pi / n)
f = np.zeros_like(x)
for j in range(n):
for k in range(n):
f[j] += x[k] * omega(j, k)
return f
def round_complex(x, decimals=0):
return np.round(np.real(x), decimals=decimals) + 1j * np.round(np.imag(x), decimals=decimals)
x = np.exp(2j * np.pi * np.arange(8) / 8)
x
f = dft(x)
f
round_complex(f)
On va faire des tests numériques, avec une version "optimisée" automatiquement grâce à numba.jit.
@jit
def dft_jit(x):
n = len(x)
pi_2j_by_n = 2j * np.pi / n
f = np.zeros_like(x)
for j in range(n):
for k in range(n):
f[j] += x[k] * np.exp(- (j * k) * pi_2j_by_n)
return f
round_complex(dft_jit(x))
Implémentation de FFT dans le module numpy¶
f2 = np.fft.fft(x)
round_complex(f2)
In this example, real input has an FFT which is Hermitian, i.e., symmetric
in the real part and anti-symmetric in the imaginary part, as described in
the numpy.fft documentation:
import matplotlib.pyplot as plt
t = np.arange(256)
sp = np.fft.fft(np.sin(t))
freq = np.fft.fftfreq(t.shape[-1])
plt.plot(freq, sp.real, freq, sp.imag)
Implémentation de la DFT par multiplication matricielle¶
On peut calculer la matrice de Vandermonde $W_N \in\mathbb{C}^{N \times N}$, associée à la racine $\omega_N = \exp(-i \frac{2\pi}{N}$ pour un signal de taille $N$, et ensuite calculer la DFT(x) comme $z = W_N x$ (produit matriciel). Cf cette page Wikipédia.
$$ f_j = \sum_{k=0}^{n-1} x_k e^{-{2\pi i \over n} jk } \qquad j = 0,\dots,n-1. $$ou en notation matricielle :
$$ \begin{array}{l} \begin{pmatrix} f_0 \\ f_1 \\ f_2 \\ \vdots \\ f_{n-1} \end{pmatrix} = \begin{pmatrix} 1 & 1 & 1 & \cdots & 1\\ 1 & w & w^2 & \cdots & w^{n-1}\\ 1 & w^2 & w^4 & \cdots & w^{2(n-1)}\\ \vdots & \vdots & \vdots & \ddots & \vdots &\\ 1 & w^{n-1} & w^{2(n-1)} & \cdots & w^{(n-1)^2} \end{pmatrix} \end{array} \begin{pmatrix} x_0 \\ x_1 \\ x_2 \\ \vdots \\ x_{n-1} \end{pmatrix} , w = e^{-\frac{2 \pi i}{n}} $$def vandermonde_fourier(n):
pi_2j_by_n = 2j * np.pi / n
omega = np.zeros((n, n), dtype=np.complex)
for j in range(n):
for k in range(n):
omega[j, k] = np.exp(- (j * k) * pi_2j_by_n)
return omega
def dft_naive_matmult(x):
n = len(x)
return vandermonde_fourier(len(x)) * np.array(x)
f3 = dft_naive_matmult(x)
round_complex(f3)
%timeit dft_naive_matmult(random_complex_vector(2**8))
Une approche plus efficace :
def vandermonde_fourier2(n):
pi_2j_by_n = 2j * np.pi / n
omega = np.zeros((n, n), dtype=np.complex)
for j in range(n):
omega[j, :] = np.exp(- (j * np.arange(n)) * pi_2j_by_n)
return omega
def dft_naive_matmult2(x):
n = len(x)
return vandermonde_fourier2(len(x)) * np.array(x)
f4 = dft_naive_matmult2(x)
round_complex(f4)
%timeit dft_naive_matmult2(random_complex_vector(2**8))
Une approche encore un peu plus efficace :
@jit
def vandermonde_fourier3(n):
pi_2j_by_n = 2j * np.pi / n
omega = np.zeros((n, n), dtype=np.complex)
for j in range(n):
omega[j, :] = np.exp(- (j * np.arange(n)) * pi_2j_by_n)
return omega
def dft_naive_matmult3(x):
n = len(x)
return vandermonde_fourier3(len(x)) * np.array(x)
f5 = dft_naive_matmult3(x)
round_complex(f5)
%timeit dft_naive_matmult3(random_complex_vector(2**8))
Implémentation manuelle de la FFT (Cooley-Tucker)¶
Comme avec notre petite implémentation de l'algorithme de Strassen, on va se restreindre aux valeurs $n = 2^p$ pour $p=\mathbb{N}$.
On va utiliser la DFT naïve pour les valeurs trop petites, comme on sait que trop d'appels récursifs ajoutent un gros surcoût (avec leaf_size=64, cela devrait donner un bon compromis).
LEAF_SIZE = 64
def fft(x, leaf_size=LEAF_SIZE):
n = len(x)
if n <= 1:
return x
elif n <= leaf_size:
return dft(x)
n_by_2 = n//2
assert n == 2 * n_by_2, "Error : only n = 2^k are accepted."
# we split the entries in 2 vectors of size n/2
# we compute the two FFT, recursively, in T(n/2)
even_fft = fft(x[0::2], leaf_size=leaf_size)
odd_fft = fft(x[1::2], leaf_size=leaf_size)
# combine the two, in O(n)
full_fft = np.zeros(n, dtype=np.complex)
omega_n = np.exp(-2j * np.pi / n)
omega_s = omega_n ** np.arange(n_by_2) # compute all the omega^j j=0..n/2
full_fft[:n_by_2] = even_fft[:] + omega_s * odd_fft[:]
full_fft[n_by_2:] = even_fft[:] - omega_s * odd_fft[:]
# so T(n) = O(n) + 2 T(n/2)
# ==> T(n) = O(n \log(n)) by master theorem!
return full_fft
Un exemple, pour vérifier notre implémentation :
f6 = fft(x, leaf_size=1)
round_complex(f6)
round_complex(fft(x, leaf_size=2))
round_complex(fft(x))
round_complex(np.fft.fft(x))
Cette implémentation semble fonctionner sans problème.
Pour les tests numériques, on peut aussi écrire une variante avec numba.jit.
@jit
def fft_jit(x, leaf_size=LEAF_SIZE):
n = len(x)
if n <= 1:
return x
elif n <= leaf_size:
return dft_jit(x)
n_by_2 = n//2
assert n == 2 * n_by_2, "Error : only n = 2^k are accepted."
# we split the entries in 2 vectors of size n/2
# we compute the two FFT, recursively, in T(n/2)
even_fft = fft_jit(x[0::2], leaf_size=leaf_size)
odd_fft = fft_jit(x[1::2], leaf_size=leaf_size)
# combine the two, in O(n)
full_fft = np.zeros(n, dtype=np.complex)
omega_n = np.exp(-2j * np.pi / n)
omega_s = omega_n ** np.arange(n_by_2) # compute all the omega^j j=0..n/2
full_fft[:n_by_2] = even_fft[:] + omega_s * odd_fft[:]
full_fft[n_by_2:] = even_fft[:] - omega_s * odd_fft[:]
# so T(n) = O(n) + 2 T(n/2)
# ==> T(n) = O(n \log(n)) by master theorem!
return full_fft
Tests avec des vecteurs aléatoires¶
On va définir des vecteurs $x\in\mathbb{C}^n$ aléatoires, distribués selon des lois normales centrées.
def random_complex_vector(n=16):
return np.random.standard_normal(size=n) + 1j * np.random.standard_normal(size=n)
x = random_complex_vector(4)
x
f1 = np.fft.fft(x)
f1
f2 = dft(x)
f2
f3 = dft_jit(x)
f3
f4 = fft(x, leaf_size=2)
f4
f5 = fft_jit(x, leaf_size=2)
f5
Maintenant quelques comparaisons, montrant que l'implémentation naïve est très mauvaise en comparaison de celle optimisée (en C) de numpy.fft, et que l'implémentation de la FFT est aussi plutôt lente :
for n in tqdm([2**4, 2**5, 2**6, 2**7, 2**8, 2**9, 2**10]):
print(f"""\nPour des vecteurs aléatoires de tailles {n}
numpy.fft.fft | dft naive | dft numba.jit | fft naive | fft jit.jit """)
x = random_complex_vector(n)
%timeit np.fft.fft(x)
assert np.all(np.isclose(np.fft.fft(x), dft(x)))
%timeit dft(x)
assert np.all(np.isclose(np.fft.fft(x), dft_jit(x)))
%timeit dft_jit(x)
assert np.all(np.isclose(np.fft.fft(x), fft(x)))
%timeit fft(x)
assert np.all(np.isclose(np.fft.fft(x), fft_jit(x)))
%timeit fft_jit(x)
round(258_000 / 9.79)
Avec une taille aussi petite que juste n=2**8=256 échantillons, la DFT naïve (en $\Theta(n^2)$) et en pure Python est déjà environ 20_000 fois plus lente que la FFT optimisée.
La DFT naïve mais compilée avec numba.jit est quant à elle 1000 plus lente.
La FFT naïve devrait être en $\mathcal{O}(n \log(n))$ et on a l'impression de le vérifier ici !
Quelle est l'influence de leaf_size ici ?
for n in tqdm([2**7, 2**8, 2**9]):
print(f"""\nPour des vecteurs aléatoires de tailles {n}
numpy.fft.fft | fft naive | fft jit.jit for different leaf_size""")
x = random_complex_vector(n)
%timeit np.fft.fft(x)
for leaf_size in [1, 8, 32, 64, 2*n]:
print(f"For leaf_size = {leaf_size}")
assert np.all(np.isclose(np.fft.fft(x), fft(x, leaf_size=leaf_size)))
%timeit fft(x)
assert np.all(np.isclose(np.fft.fft(x), fft_jit(x, leaf_size=leaf_size)))
%timeit fft_jit(x)
The influence is hard to see, but this is very counter intuitive.
Conclusion¶
C'est bon pour aujourd'hui !